Interpretation of the outputs of PREDIPATH script
Date: 07/12/19; time: 14:02.
Your text results:
Input file: myGenome(45HUQ.fasta
Genus: Ralstonia
Classes:
NPP = Not plant-pathogenic
PP = Plant-pathogenic
Search of specific k-mers
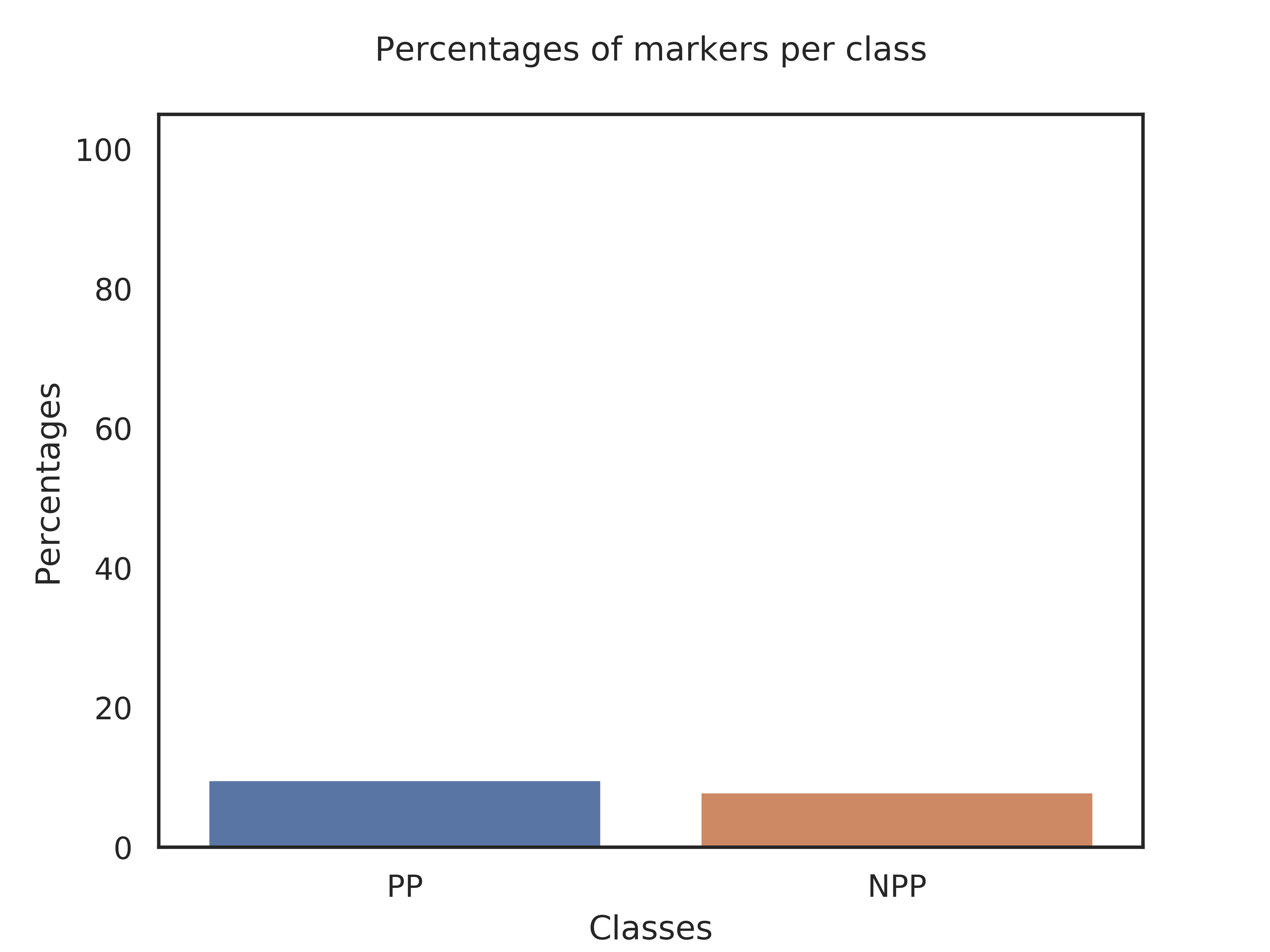
Classes,Kmers_per_class,Query_genome,Percent
PP,5167,500,9.68%
NPP,1262,100,7.92%
Search of specific genes
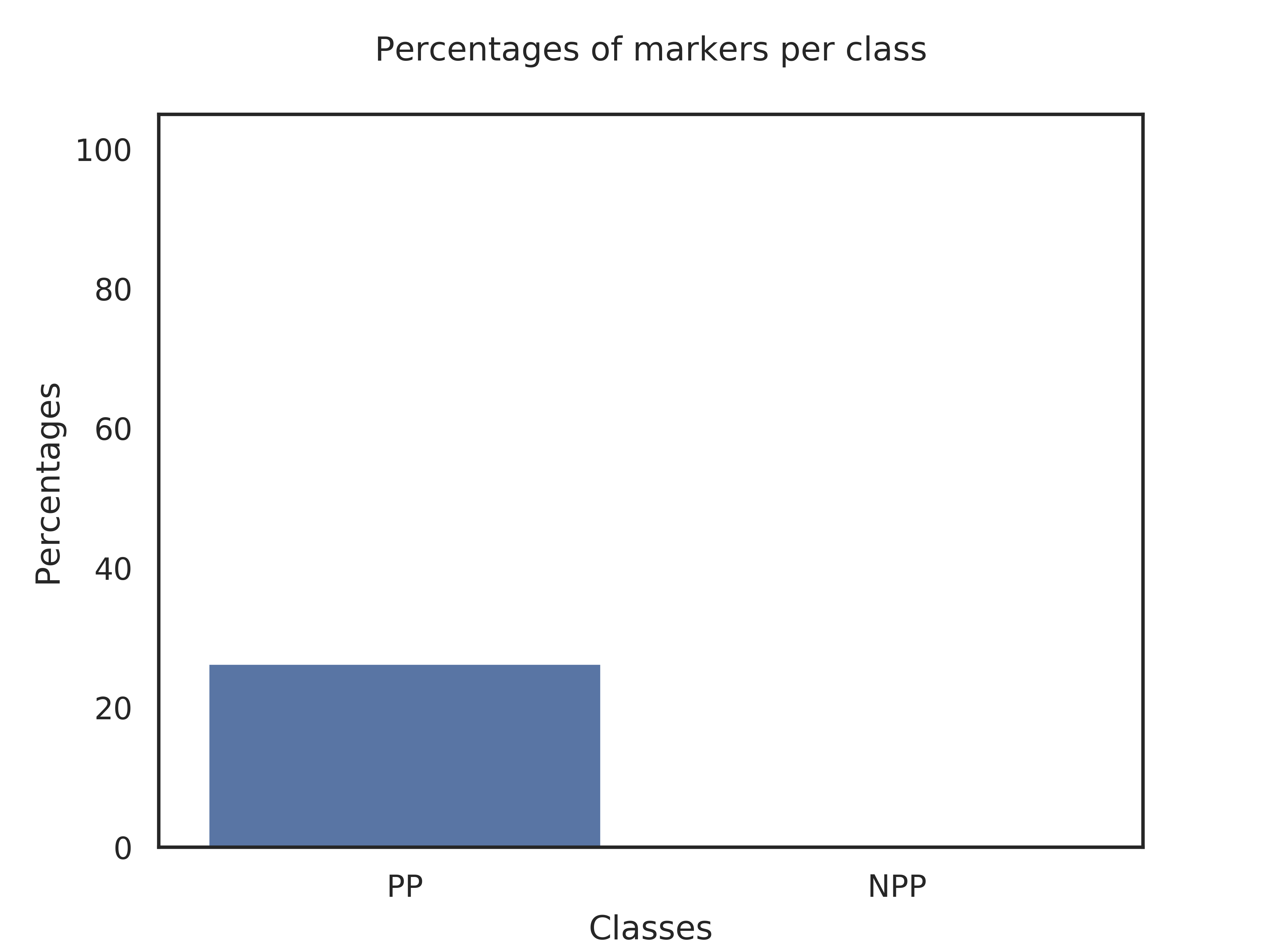
Classes,Genes_per_class,Query_genome,Percent
PP,19,5,26.32%
NPP,1,0,0.0%
Informations
Your genome was read in the file: myGenome(45HUQ.fasta
You are working with genus: Ralstonia
2 classes are defined:
- class 1 short name: NPP long name: Not plant-pathogenic
- class 2 short name: PP long name: Plant-pathogenic
Explanation of results for the block: Search of specific k-mers
Class PP is defined by 5167 markers. Your query genome has 500 of these makers, which represents 9.68% of the markers.
Class NPP is defined by 1262 markers. Your query genome has 100 of these makers, which represents 7.92% of the markers.
Therefore, your genome is difficult to classify as belonging to class PP or NPP. We suggest to check if your genome really belongs to the genus Ralstonia..
Explanation of results for the block: Search of specific genes
Class PP is defined by 19 markers. Your query genome has 5 of these makers, which represents 26.32% of the markers.
Class NPP is defined by 1 markers. Your query genome has 0 of these makers, which represents 0.0% of the markers.
Therefore, your genome is difficult to classify as belonging to class PP or NPP. We suggest to check if your genome really belongs to the genus Ralstonia..