Interpretation of the outputs of PREDIPATH script
Date: 07/12/19; time: 14:02.
Your text results:
Input file: (unknown)
Genus: Ralstonia
Classes:
NPP = Not plant-pathogenic
PP = Plant-pathogenic
Search of specific k-mers
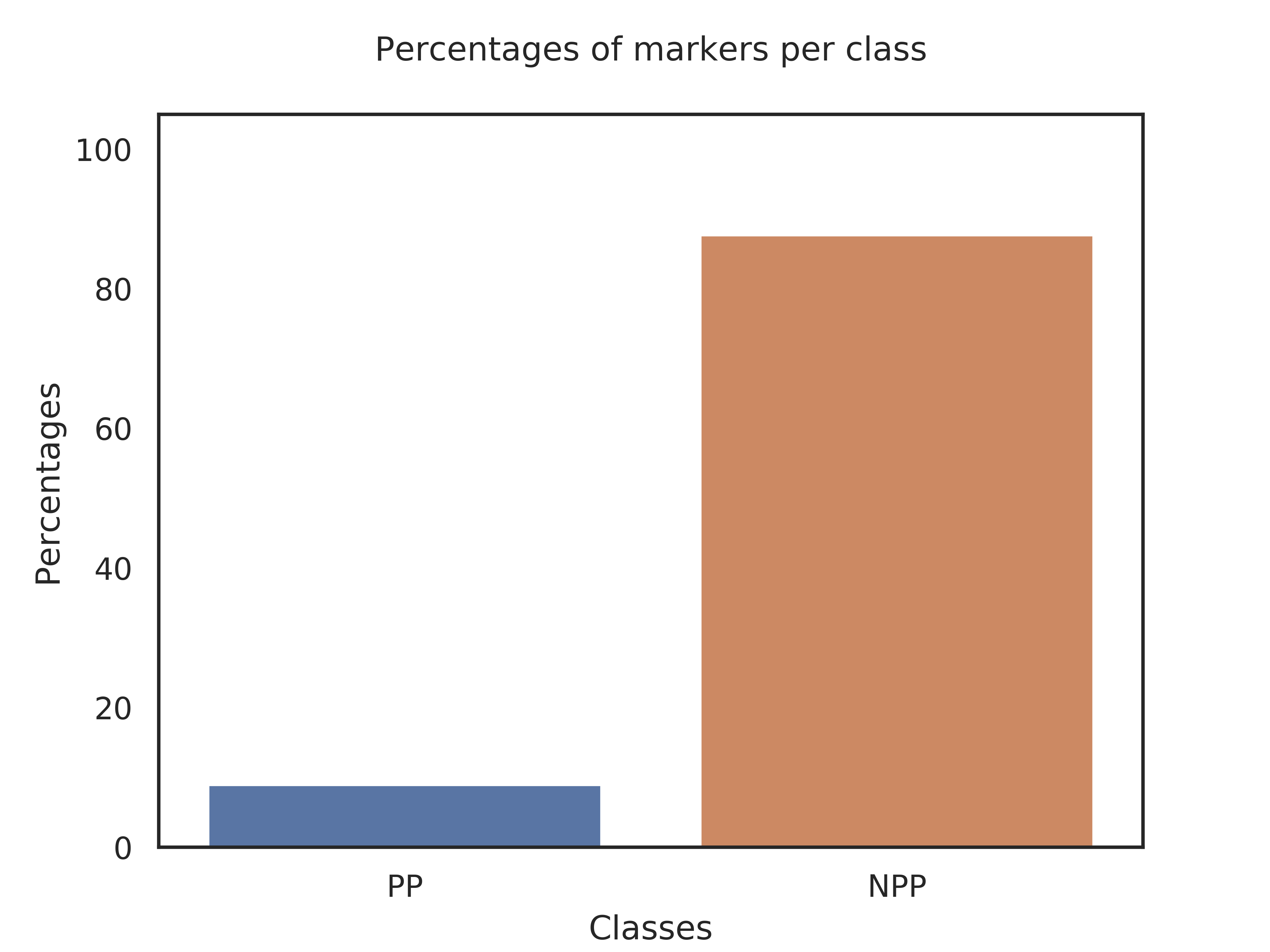
Classes,Kmers_per_class,Query_genome,Percent
PP,5167,463,8.96%
NPP,1262,1107,87.72%
Search of specific genes
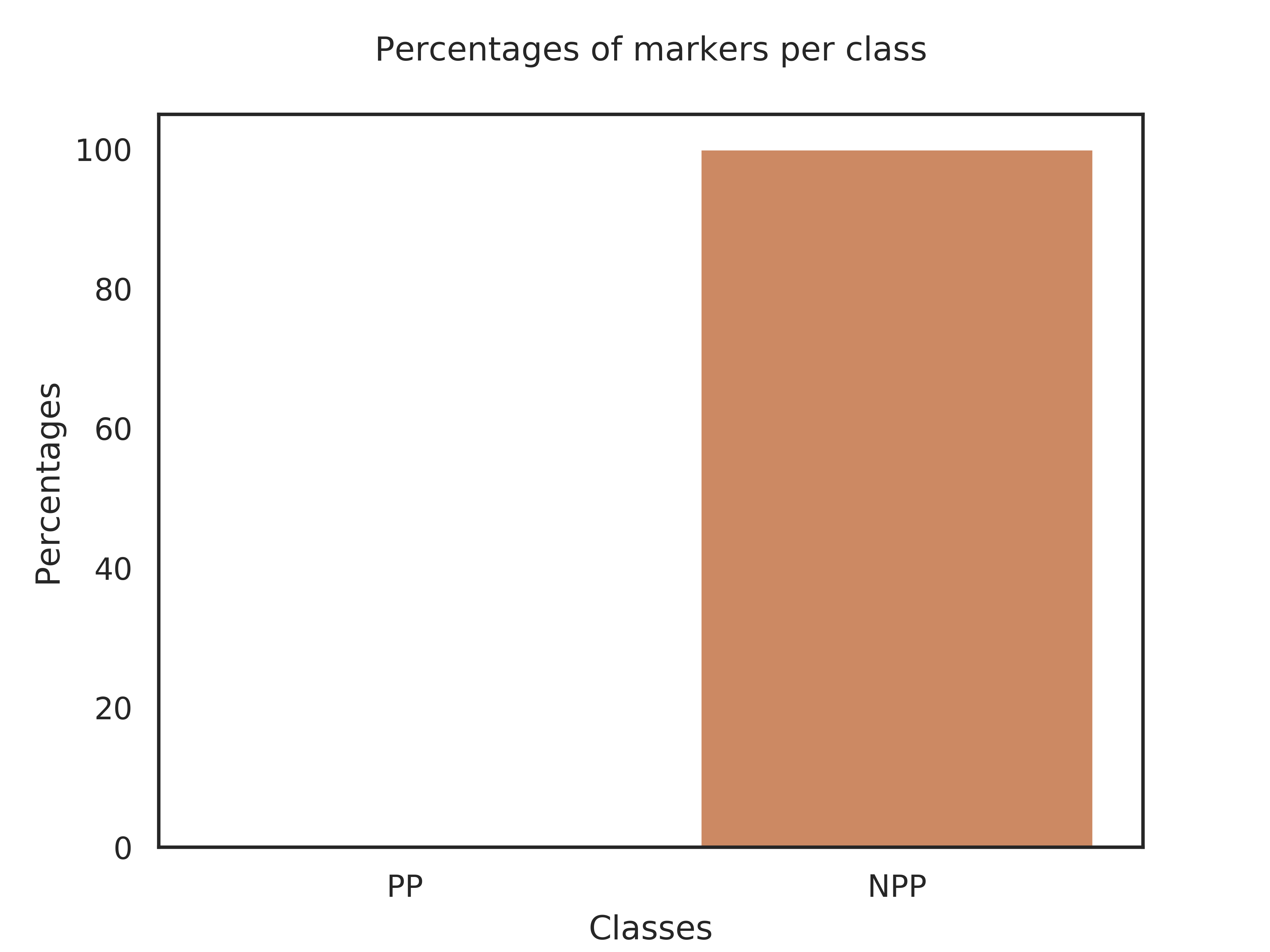
Classes,Genes_per_class,Query_genome,Percent
PP,19,0,0.00%
NPP,1,1,100.0%
Informations
Your genome was read in the file: (unknown)
You are working with genus: Ralstonia
2 classes are defined:
- class 1 short name: NPP long name: Not plant-pathogenic
- class 2 short name: PP long name: Plant-pathogenic
Explanation of results for the block: Search of specific k-mers
Class PP is defined by 5167 markers. Your query genome has 463 of these makers, which represents 8.96% of the markers.
Class NPP is defined by 1262 markers. Your query genome has 1107 of these makers, which represents 87.72% of the markers.
Therefore, your genome belongs more likely to class NPP.
Explanation of results for the block: Search of specific genes
Class PP is defined by 19 markers. Your query genome has 0 of these makers, which represents 0.00% of the markers.
Class NPP is defined by 1 markers. Your query genome has 1 of these makers, which represents 100.0% of the markers.
Therefore, your genome belongs probably to class NPP.